Advanced Drug Design with HPC
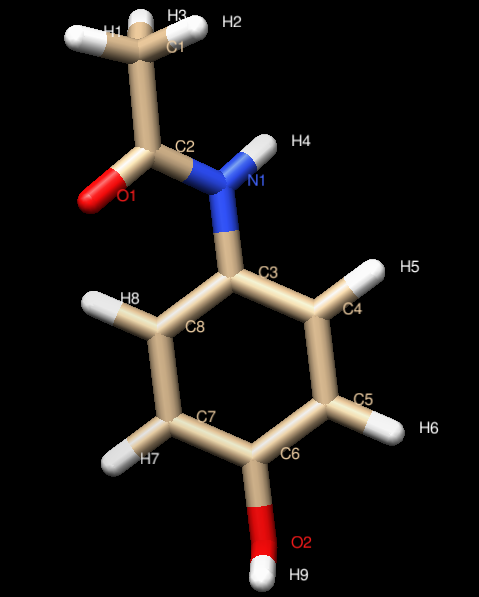
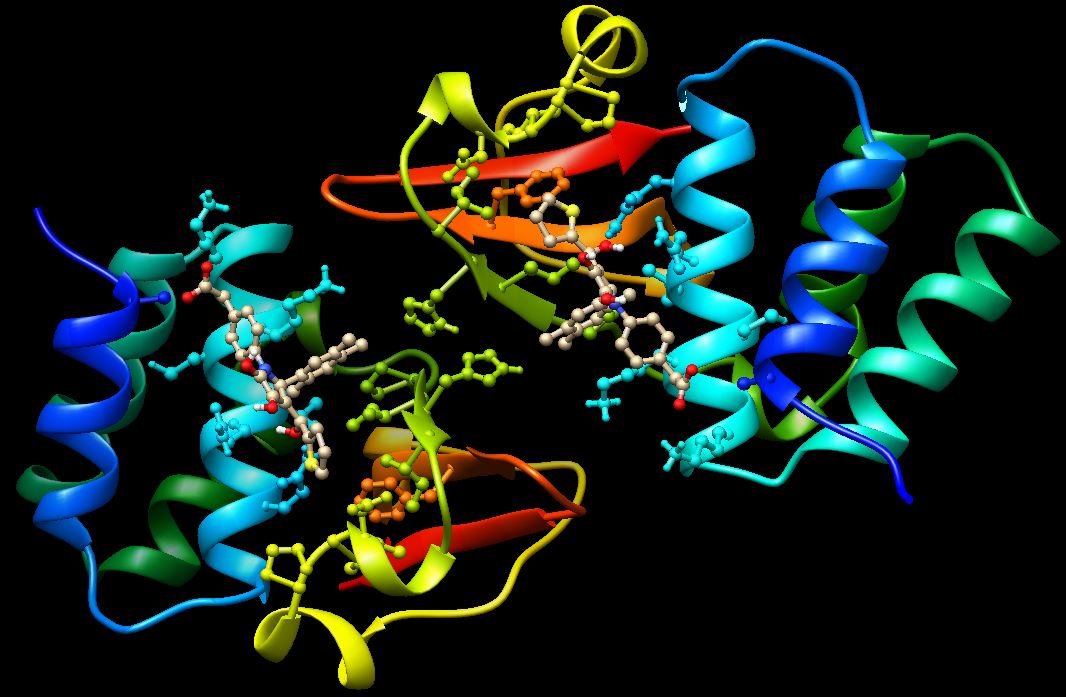
The advent of high performance cloud (HPC) computing, genomics, proteomics, bioinformatics and efficient technologies like virtual screening, in silico screening, Molecular Docking, ADMET screening and Structure-based drug design have revolutionized the way in which drugs are developed. In this training, partcipants will learn the science involved in disease target identification, virtual screening techniques, methods used in in-silico generation of ligands, protein optimization & energy minimization, molecular Docking, creation of Grid Paramater & Dock Parameter files, and running the Docking Algorithm.
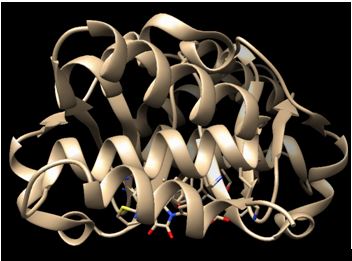
They will also learn how to select potent inhibitors on the basis of binding energies and Lipinski�s Rule of 5, how to look for H-bond between ligand and active site of the residue of protein as well as aspects of drug Likeness,ADME & Toxicity. Practical hands-on training will encompass training on the use of AWS EC2, software such as MarvinSketch, UCSF Chimera, AutoDock Tools, Open Babel, and SPDV among others.
Continue Reading »
| Dates: Every third week of the month |
| Duration: 5 days. |
| Venue: Nairobi, Kenya. |
| PREREQUISITES |
This training is ideal for people in biological, agricultural, pharma, and biotechnology industries who wish to gain skills on how to discover, design, and optimize herbal-based compounds using in silico methods. |
| FEES |
- Kshs. 30,000/- (Inclusive of VAT 16%) for East African residents. Non-East African students pay 25% more on all charges.Fee covers tuition, training material, certificate, drinks and meals only.
- Fees are payable strictly in advance (at least 4 working days before the starting date.
- Groups with a minimum of 5 people will enjoy a 10% discount and early bird registration (at least 2 weeks before the start) will be offered a 5% discount
- There will be a 10% administrative charge for cancellations received in writing up to 20 working days before the start of the course. No refunds will be made for cancellations received within 20 working days of the course start date or for the inability to attend the course for whatever reasons. KIBs is not liable for non-attendance due to travel disruptions, health problems or any other reason that might lead to a delegate not being able to attend the course. Substitutions may, of course, be made at any time, providing you inform us in writing.
- Fee is payable by bankers cheque or cash deposit to: Fee is payable by bankers cheque or MPESA to: Bioinformatics Institute of Kenya. A/C No. 01192845321700, Cooperative Bank, Co-op House Branch, Nairobi, Kenya. SWIFT Code KCOOKENAXXX. Branch Code: XXX. MPESA PAY BILL: Business number: 400222. Account number: 1679821#HERB. PAYPAL: [email protected].
|
| REGISTRATION |
To register, please download the booking form fill it and send it to us through:
Email: [email protected]. |
| TOOLS AND SOFTWARE |
| A comprehensive list of tools and software is available in our Resources page |
| Course material including a comprehensive training manual and a CD with software |
| TRAINING PROGRAM |
- DAY 1: Introduction to in-silico-based drug discovery and design and its place in the drug pipeline
- DAY 1: Target identification, obtaining crystallographic protein-ligand complexes from RCSB PDB, and homology modelling of protein targets
- DAY 1: Obtaining large ligand subsets using Wget from herbal databases
- DAY 1: 1D/2D filtering of ligands using lead-likeness and Lipinski’s rule of five
- DAY 2: Preparation of coordinate files using Chimera 1.9
- DAY 2: Energy minimization using the Amber force field/CHARMM
- DAY 2: Protein-ligand re-docking and docking using the AutoDock Tools/Vina
- DAY 2: Analysis of docking results
- DAY 3: Identification of ligand binding sites through blind docking
- DAY 3: Virtual screening using PyRx and Raccoon
- DAY 3: Analysis of virtual screening results using FOX and the summarize_results.py script
- DAY 4: Ligand-based drug design and pharmacophore building
- DAY 5: Molecular dynamics using Gromacs
- DAY 5: In-silico based ADME-tox
- DAY 5: AWARD OF CERTIFICATES
|



 Video/Audio Resources
Video/Audio Resources